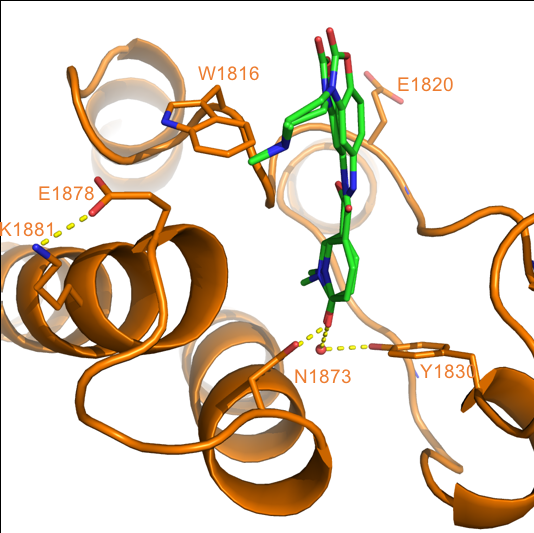
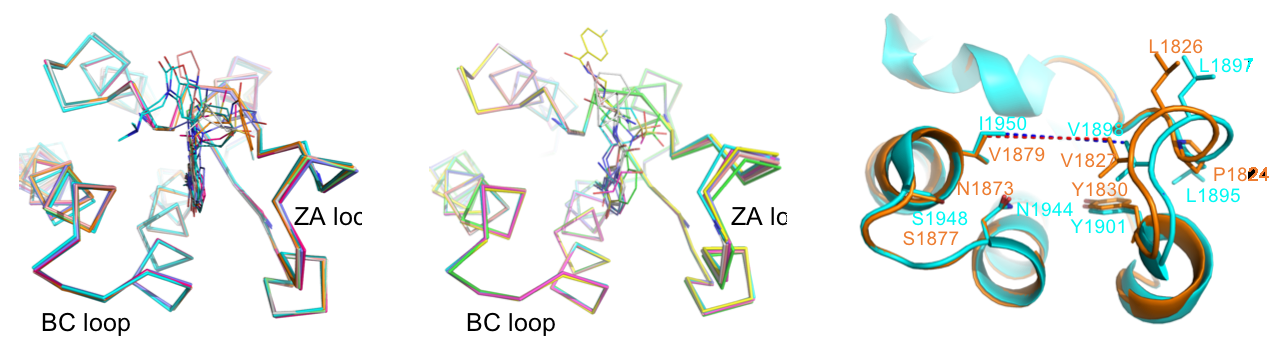
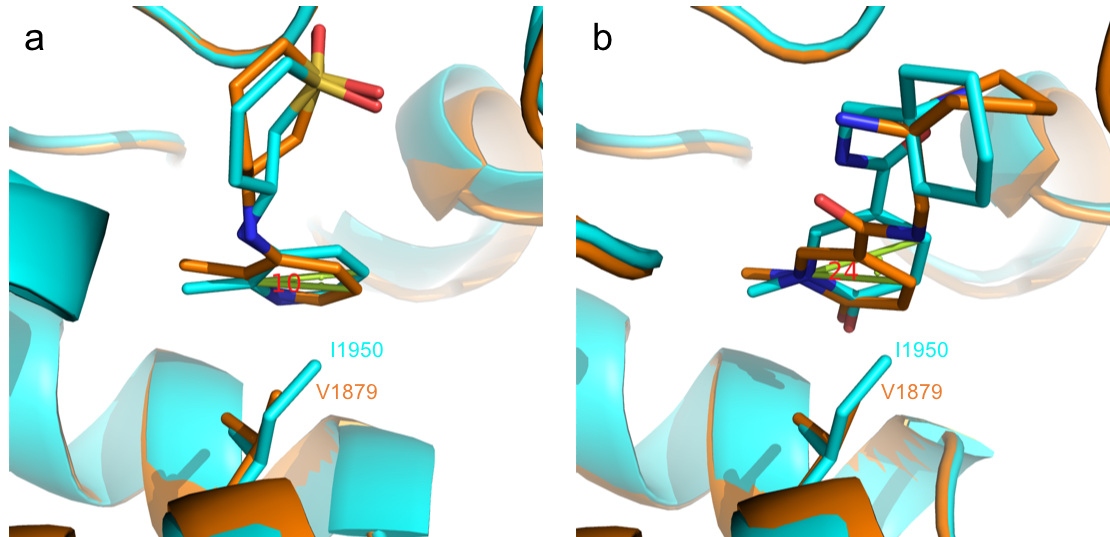
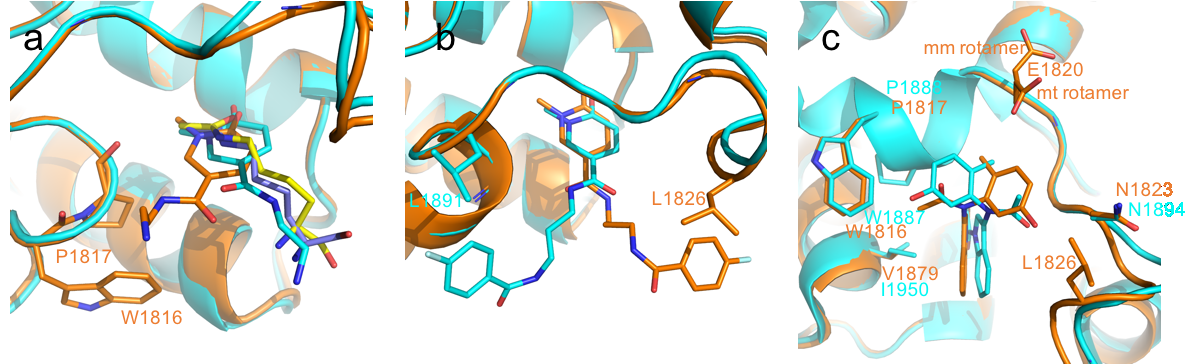
Content of the page
Stats from Google Scholar
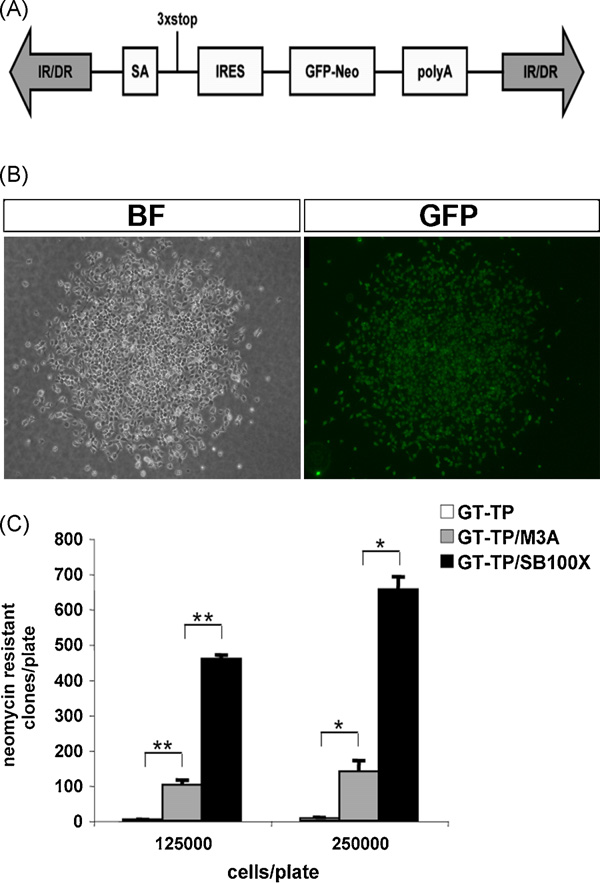
|
Citation Indices |
All | Since 2018 |
| Citations | 919 | 534 |
| h-index | 15 | 13 |
| i-10 index | 20 | 16 |
updated November 19th, 2023
My Orcid ID and QR

Peer-reviewed articles
26.
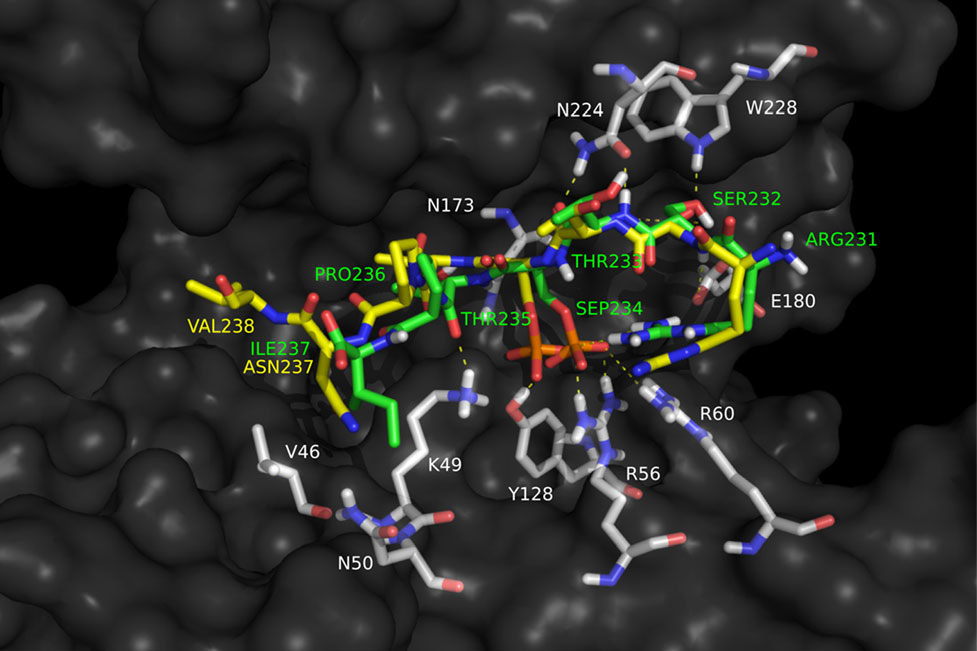
Spiliotopoulos D, Koelbert C, Audebert M, Barisch I, Bellet D, Constans M, Czich A, Finot F, Gervais V, Khoury L, Kirchnawy C, Kitamoto S, Le Tesson A, Malesic L, Matsuyama R, Mayrhofer E, Mouche I, Preikschat B, Prielinger L, Rainer B, Roblin C, Wäse K
2024. Mutat Res, 893:503718. DOI: 10.1016/j.mrgentox.2023.503718.
25.
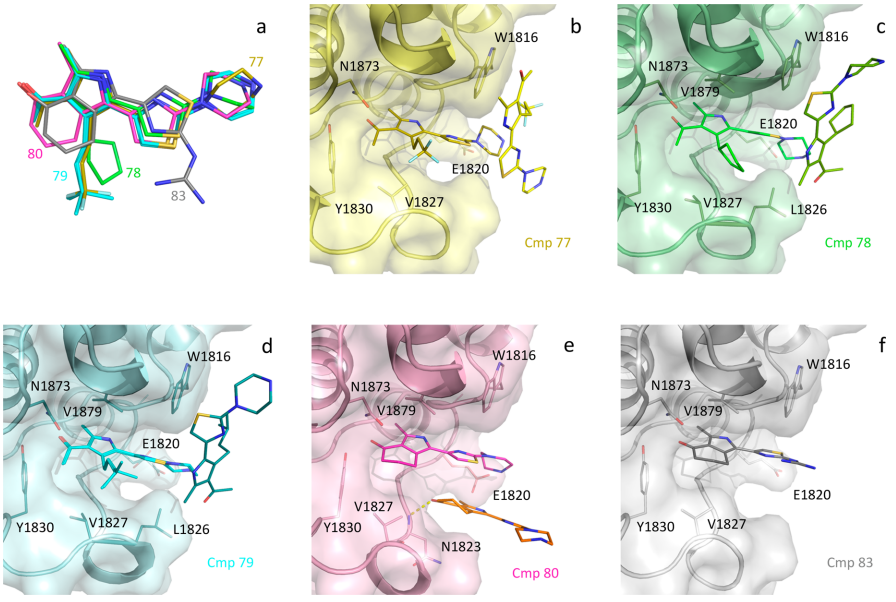
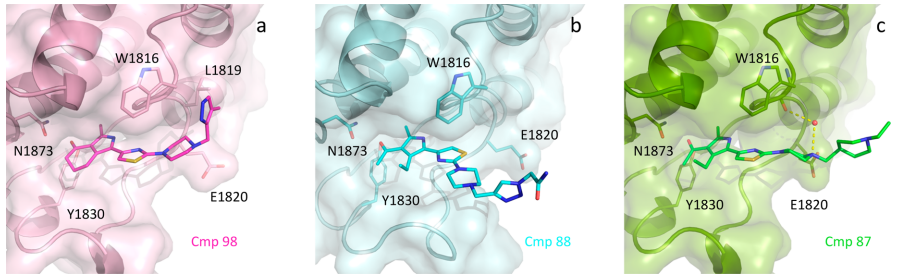
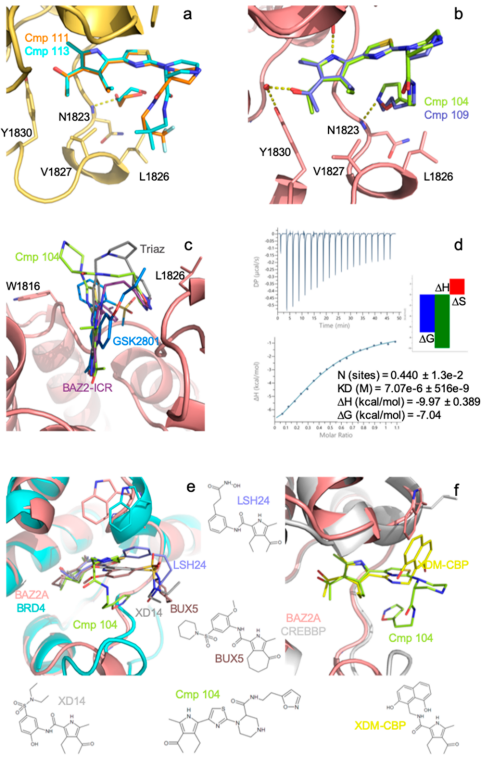
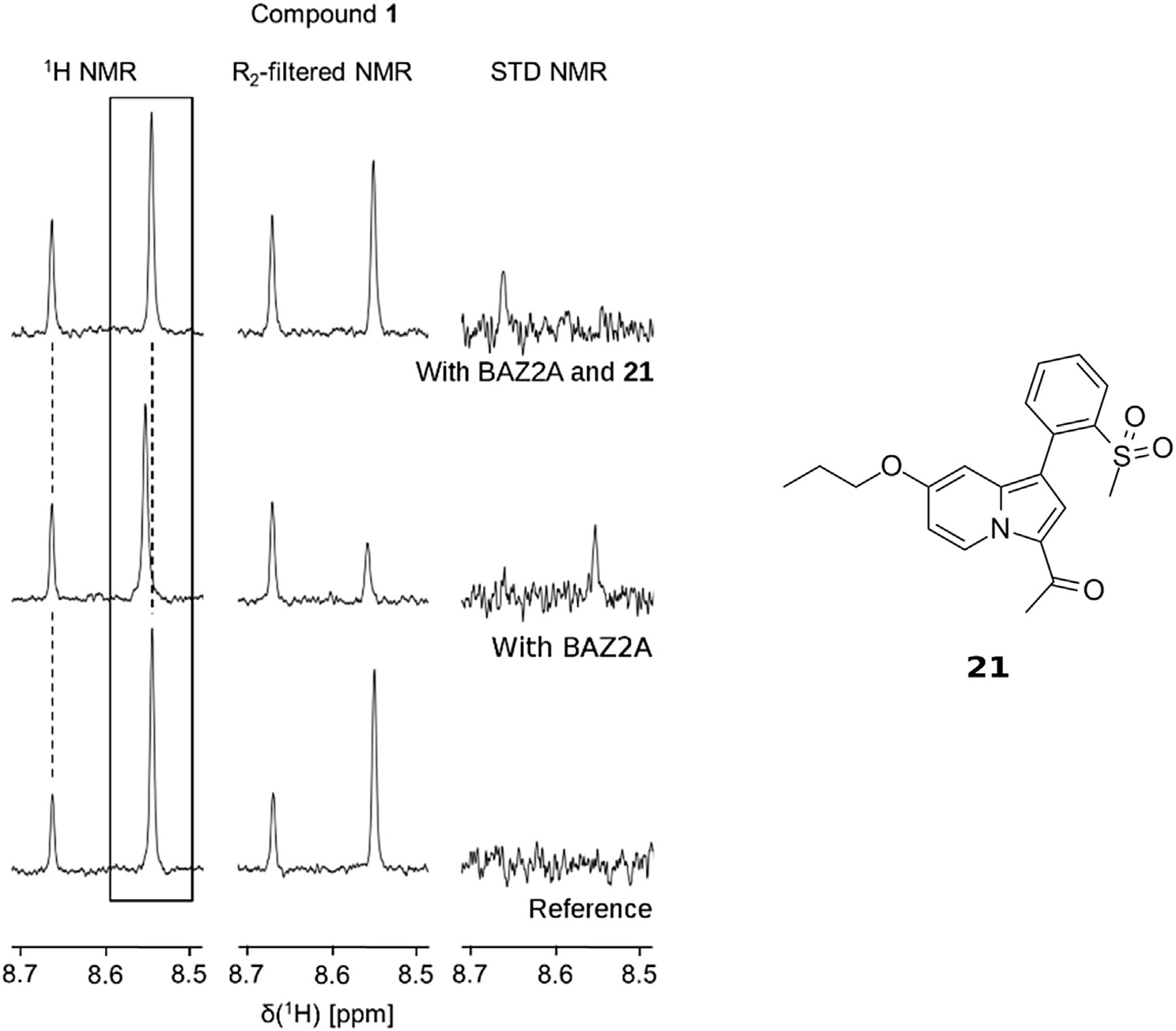
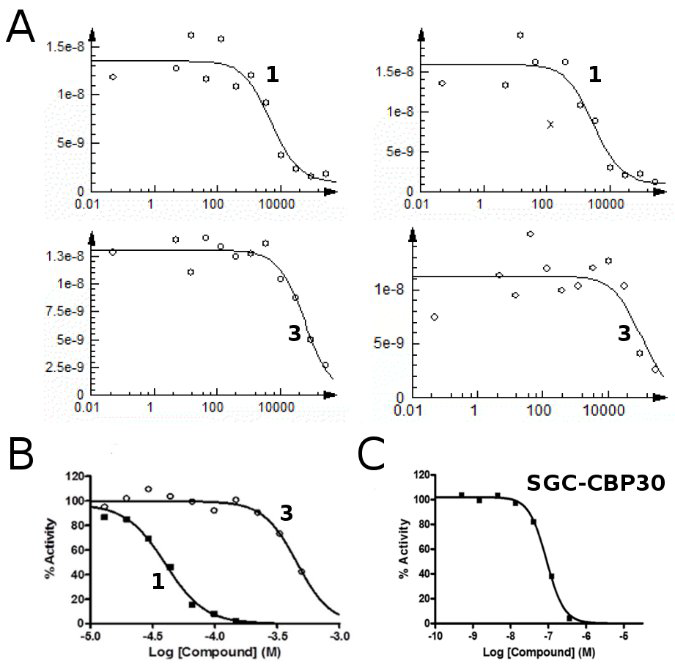
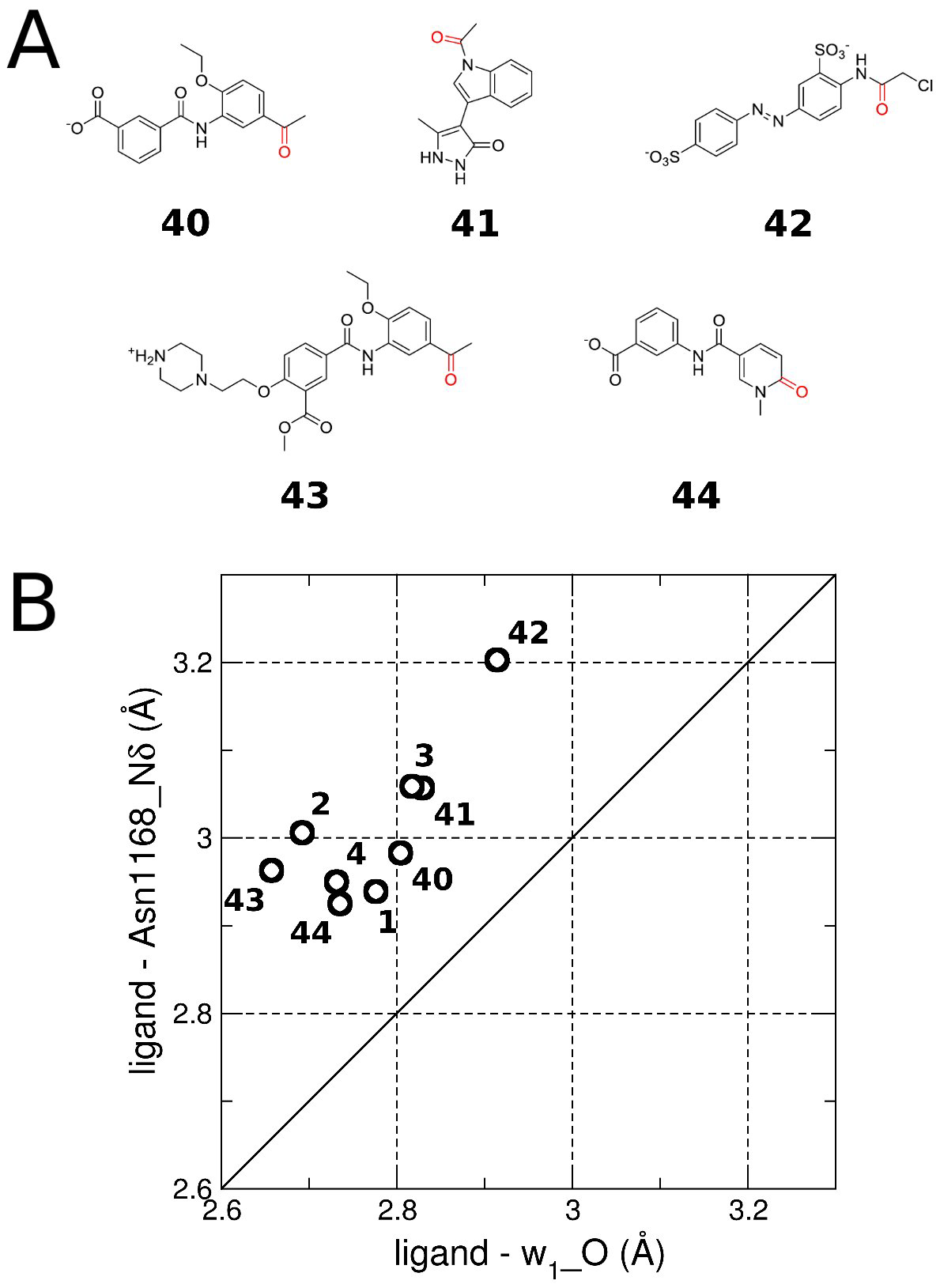
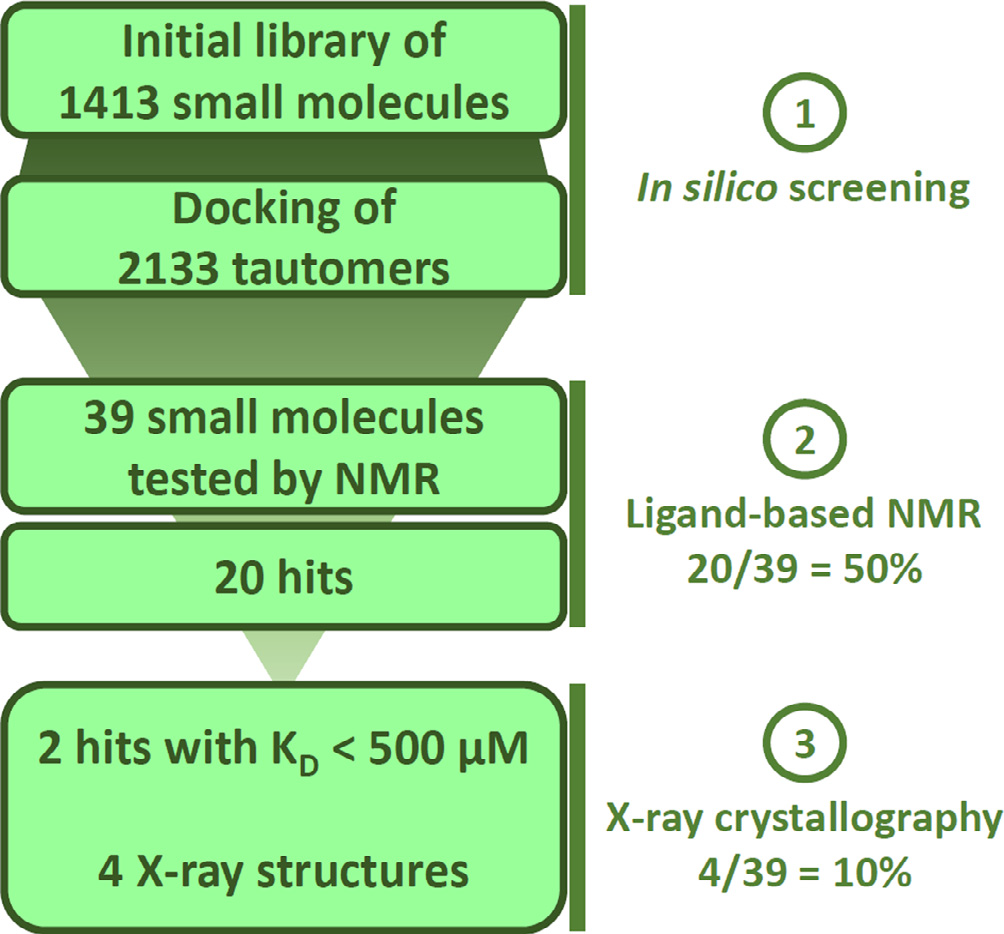
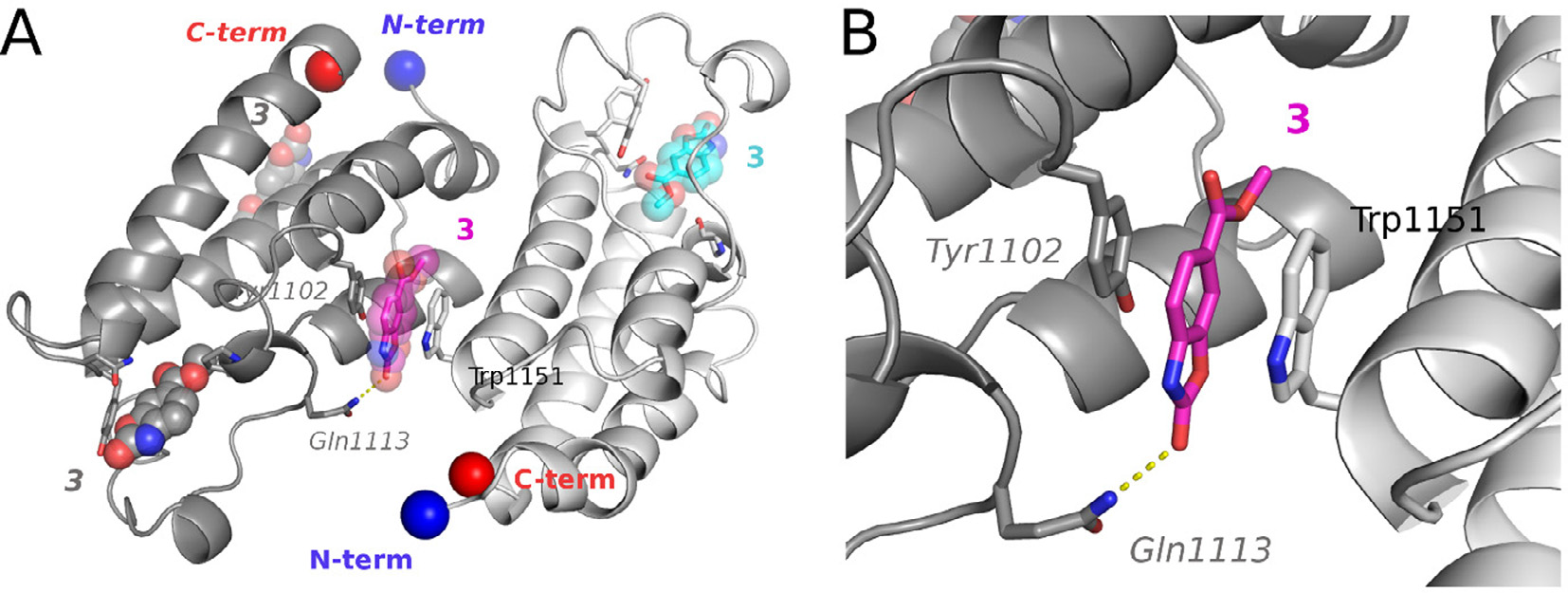
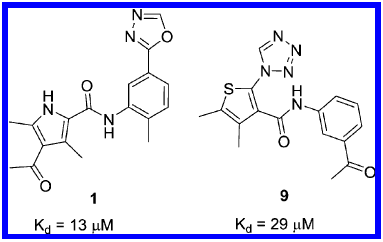
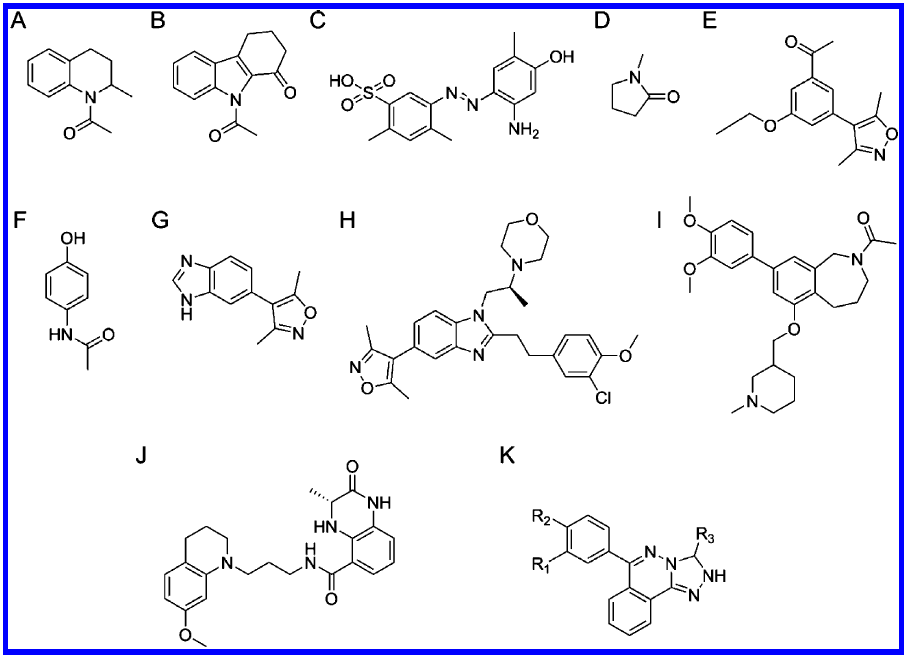
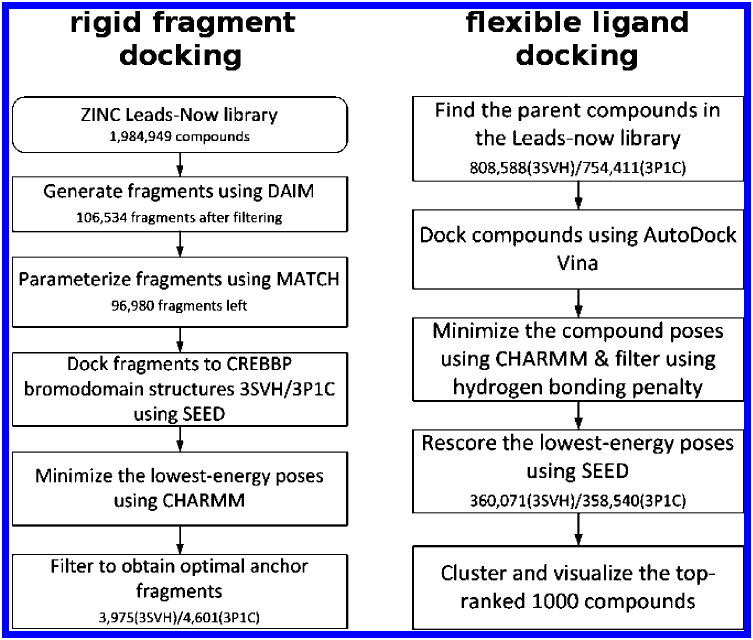
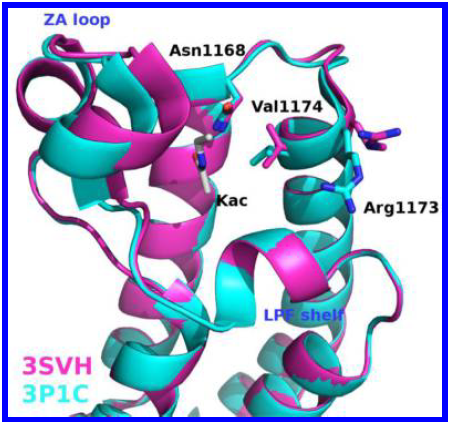
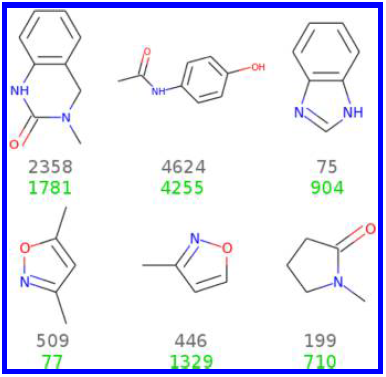
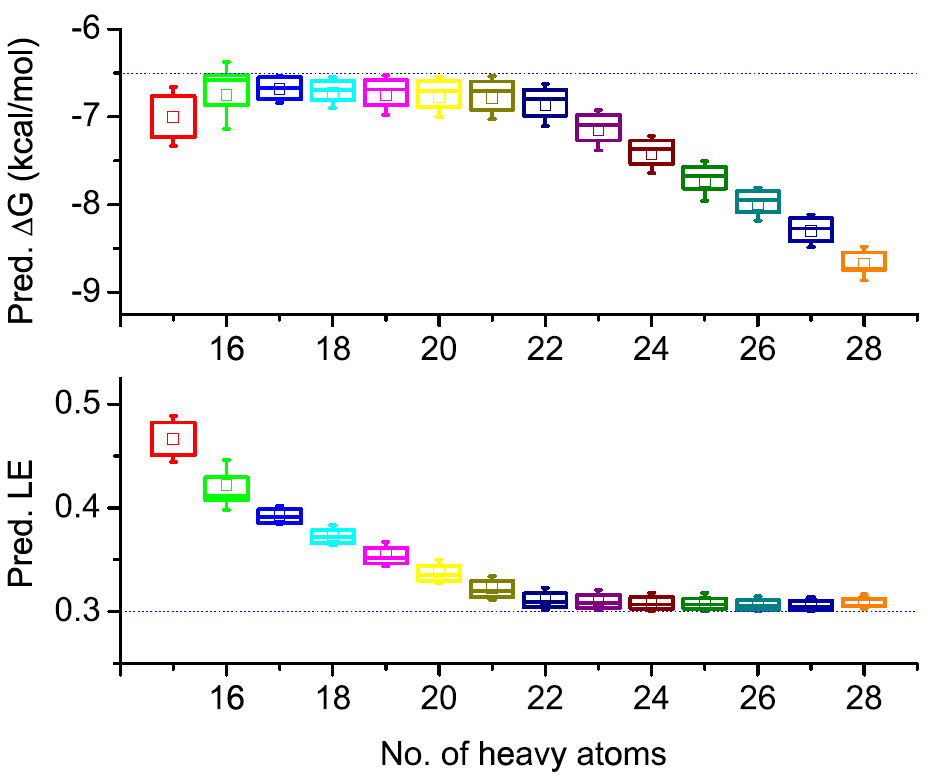
Identification of a BAZ2A-bromodomain hit compound by fragment growing
Dalle Vedove A, Cazzanelli G, Batiste L, Marchand J-R, Spiliotopoulos D, Corsi J, D'Agostino V, Caflisch A, Lolli G
2022. ACS Med Chem Lett, 13(9):1434-43. DOI: 10.1021/acsmedchemlett.2c00173.
24.
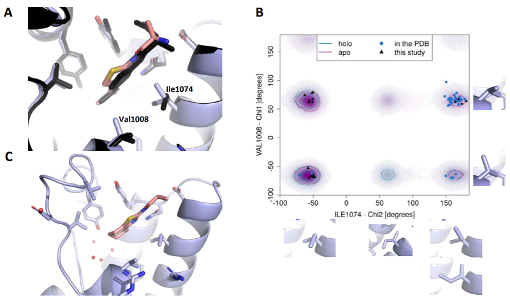
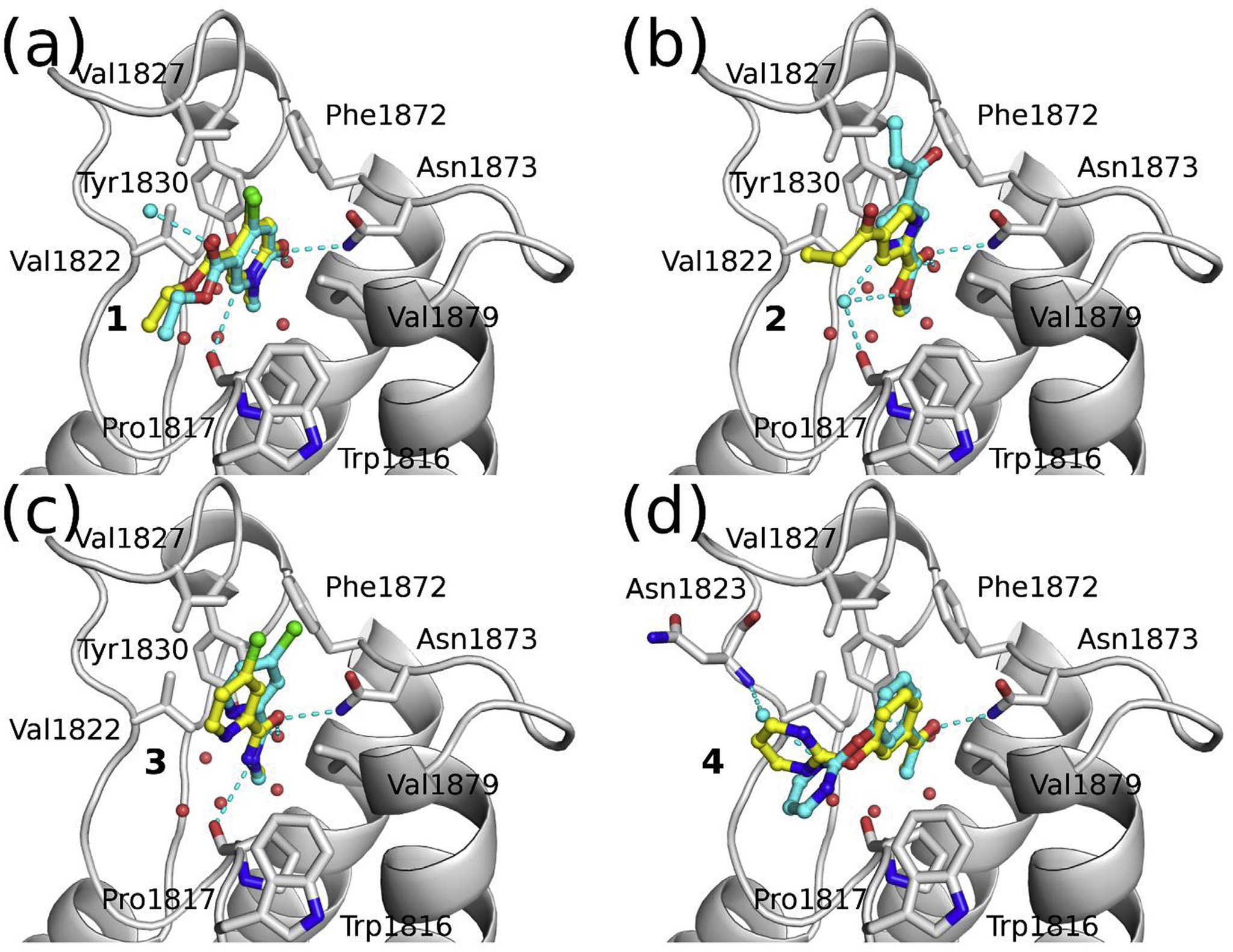
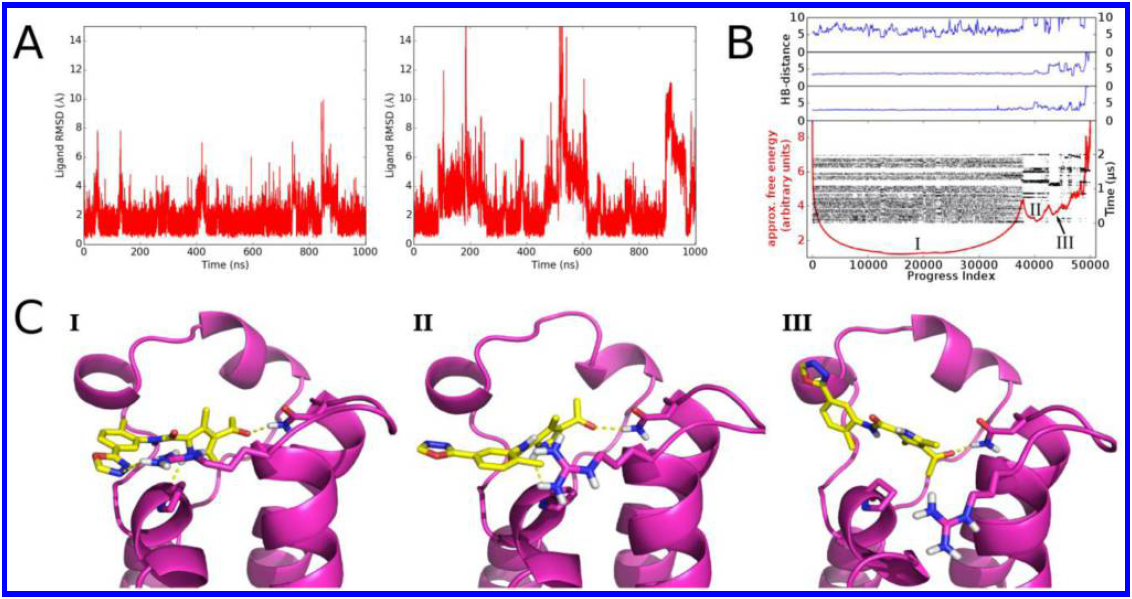
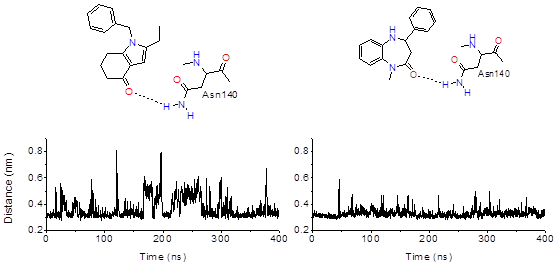
Hitting a moving target: simulation and crystallography study of ATAD2 bromodomain blockers
Dolbois A, Batiste L, Wiedmer L, Dong J, Brütsch M, Huang D, Deerain N, Spiliotopoulos D, Cheng-Sánchez I, Laul E, Nevado C, Sledz P, Caflisch A
2020. ACS Med Chem Lett, 11(8):1573–80. DOI: 10.1021/acsmedchemlett.0c00080.
23.
Spiliotopoulos D, Koelbert C
2020. Mutat Res, 856–857:503218. DOI: 10.1016/j.mrgentox.2020.503218.
22.
Understanding the mechanism of action of pyrrolo[3,2-b]quinoxaline-derivatives as kinase inhibitors
Unzue A, Jessen-Trefzer C, Spiliotopoulos D, Gaudio E, Tarantelli C, Dong J, Zhao H, Pachmayr J, Zahler S, Bernasconi E, Sartori G, Cascione L, Bertoni F, Sledz P, Caflisch A, Nevado C
2020. RCS Med Chem, 11:665–75. DOI: 10.1039/D0MD00049C.
21.
Ligand retargeting by binding site analogy
Wiedmer L, Schärer C, Spiliotopoulos D, Hürzeler M, Sledz P, Caflisch A
2019. Eur J Med
Chem, 175:107–13. DOI: 10.1016/j.ejmech.2019.04.037.
20.
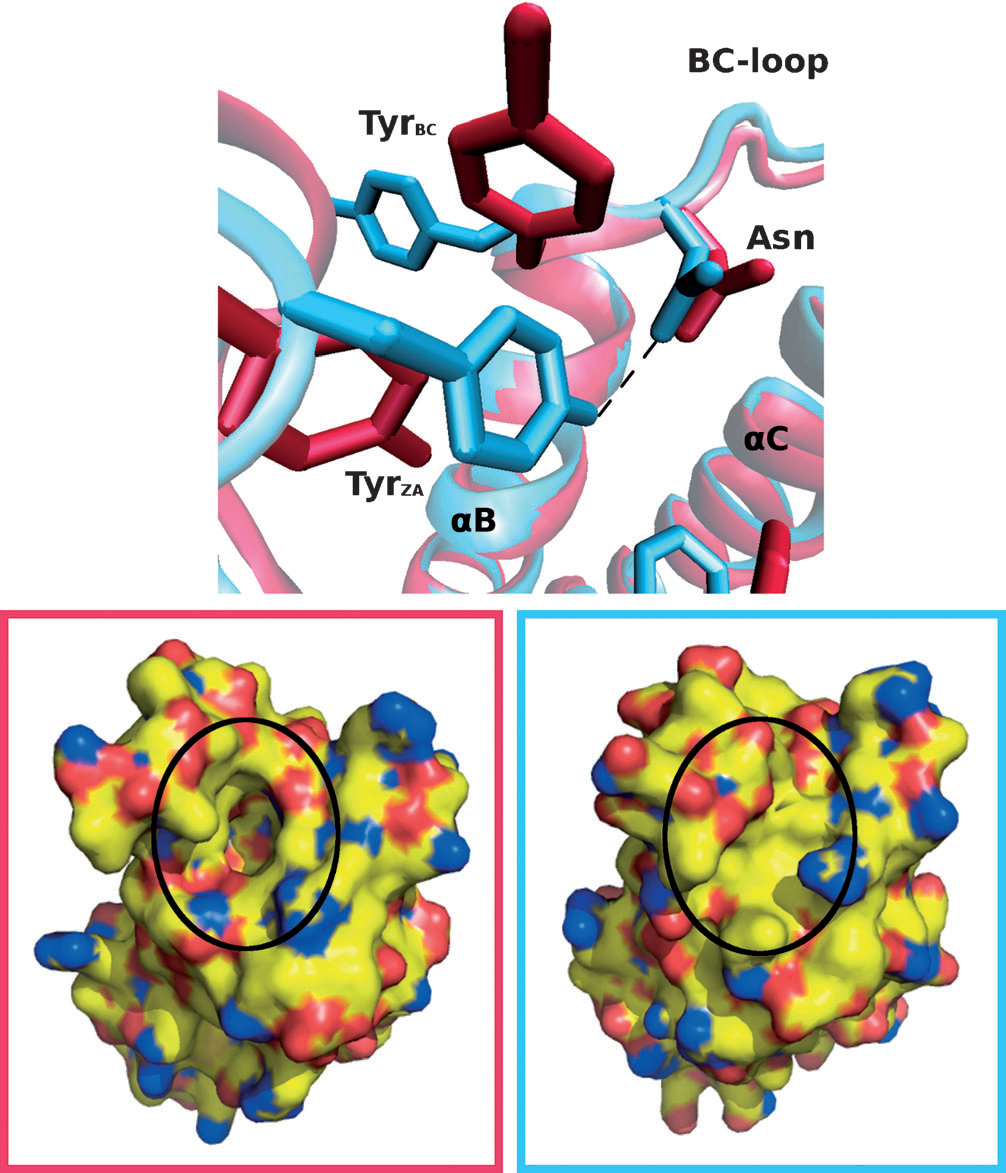
Structural analysis of small molecule binding to the BAZ2A and BAZ2B bromodomains
Dalle Vedove A, Spiliotopoulos D, D'Agostino V, Marchand, J-R, Unzue A, Nevado C, Lolli G, Caflisch A
2018. ChemMedChem, 13(14):1479–87. DOI: 10.1002/cmdc.201800234.
19.
Chemical space expansion of bromodomain ligands guided by in silico virtual couplings (autocouple)
Batiste L, Unzue A, Dolbois A, Hassler F, Wang X, Deerain N, Zhu J, Spiliotopoulos D, Nevado C, Caflisch A
2018. ACS Cent Sci, 4(2):180–8. DOI: 10.1021/acscentsci.7b00401.
18.
Discovery of BAZ2A bromodomain ligands
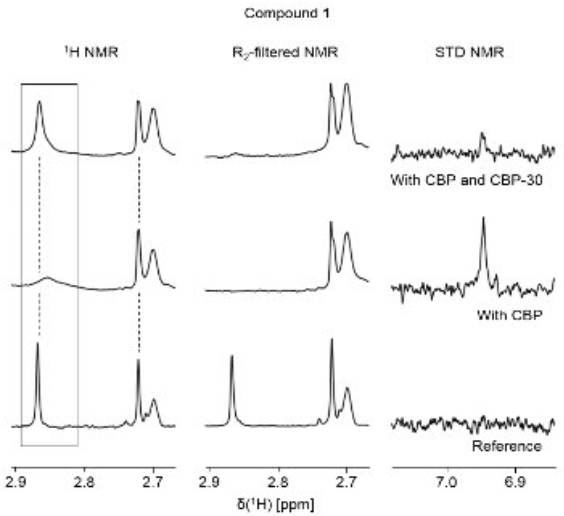
Spiliotopoulos D, Wamhoff EC, Lolli G, Rademacher C, Caflisch A
2017. Eur J Med Chem, 139:564–72. DOI: 10.1016/j.ejmech.2017.08.028.
17.
Virtual screen to NMR (VS2NMR): Discovery of fragment hits for the CBP bromodomain
Spiliotopoulos D, Zhu J, Wamhoff EC, Deerain N, Marchand J-R, Aretz J, Rademacher C, Caflisch A
2017. Bioorg Med Chem Lett, 27(11):2472–8. DOI: 10.1016/j.bmcl.2017.04.001.
16.
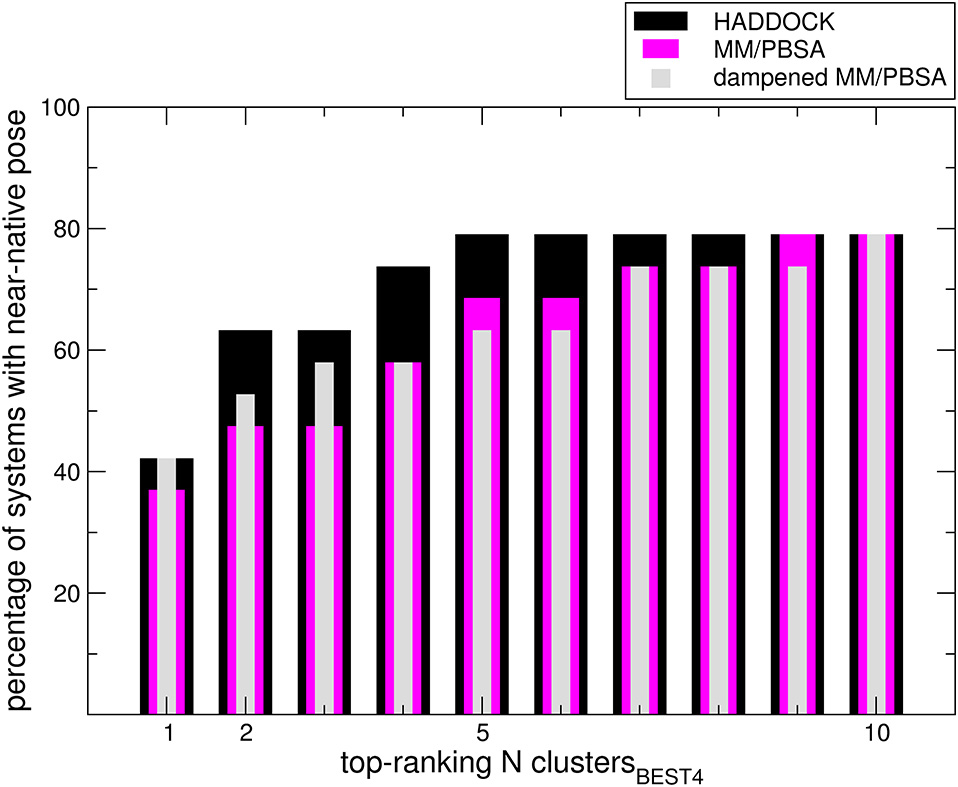
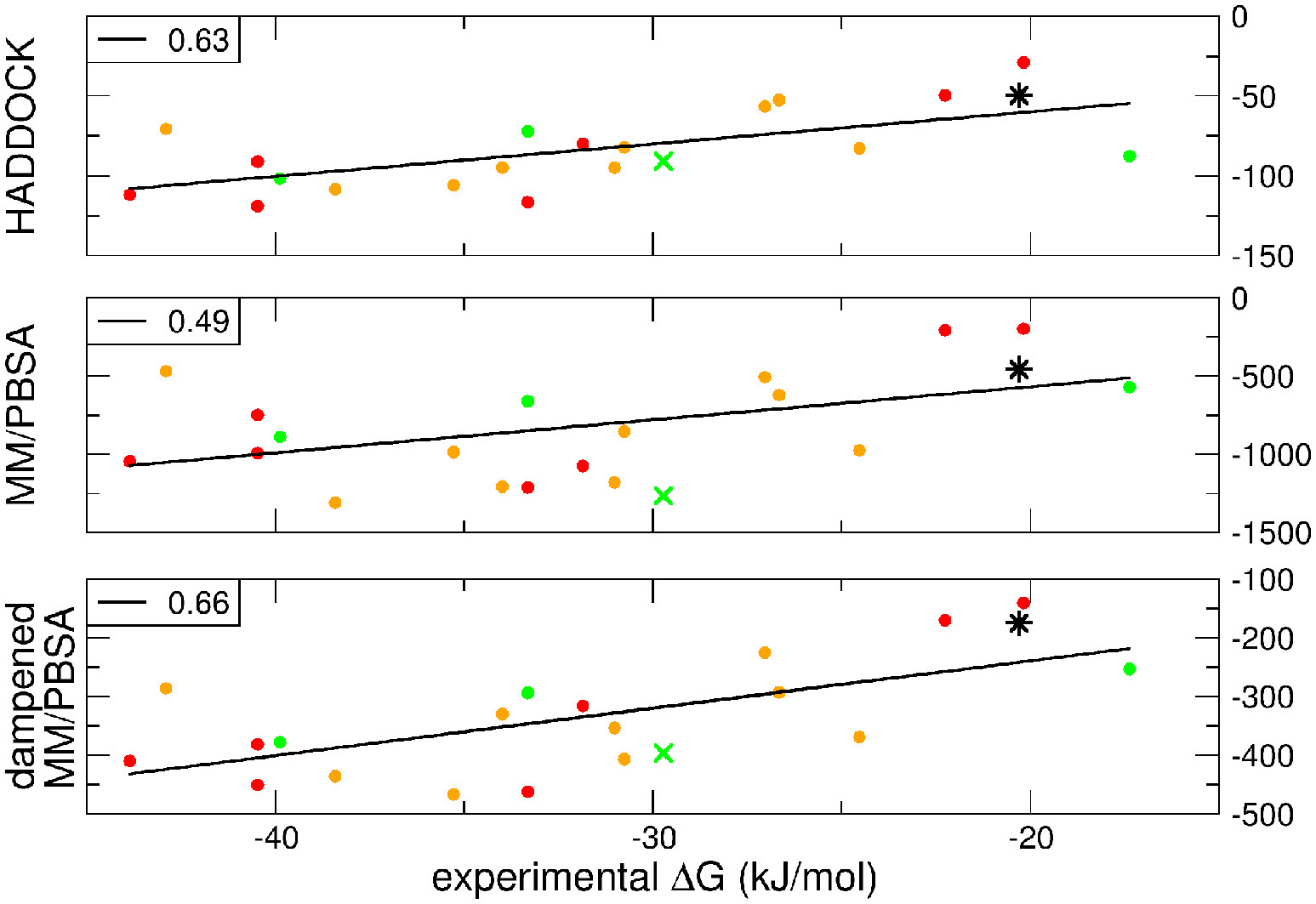
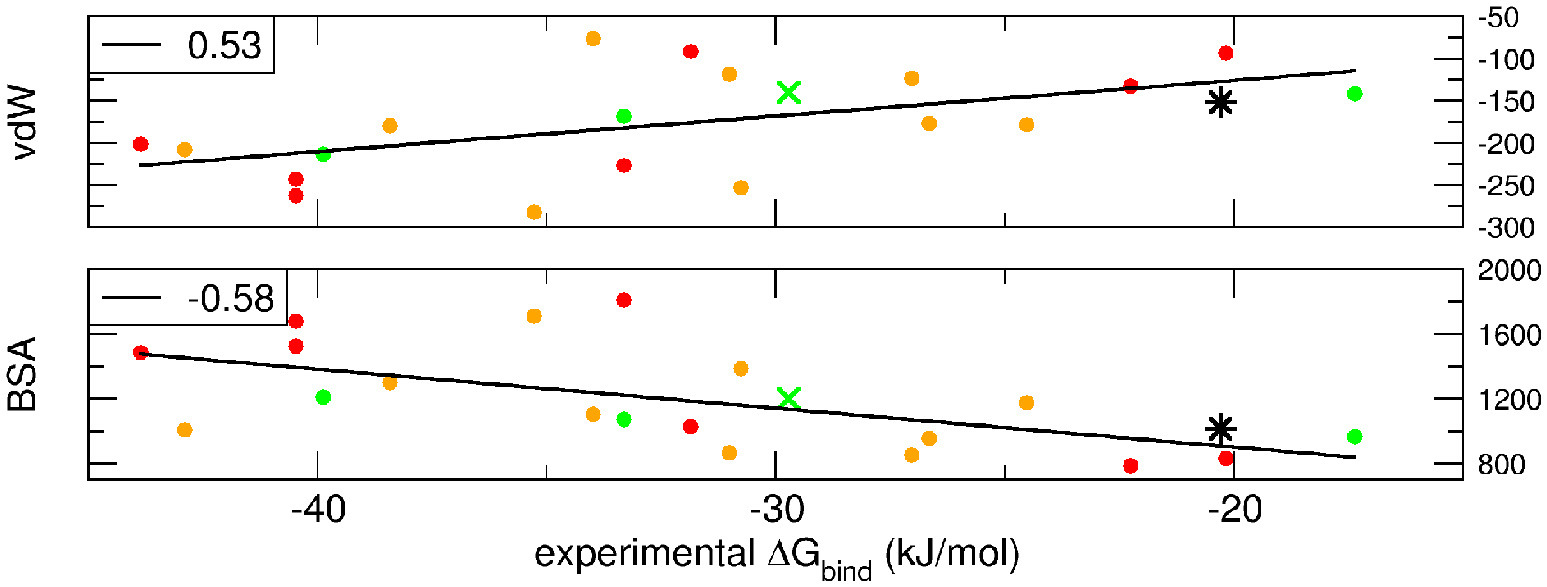
dMM-PBSA: A new HADDOCK scoring function for protein-peptide docking
Spiliotopoulos D, Kastritis PL, Melquiond A, Bonvin AMJJ, Musco G, Rocchia W, Spitaleri A
2016. Front Mol Biosci, 3:46. DOI: 10.3389/fmolb.2016.00046.
15.
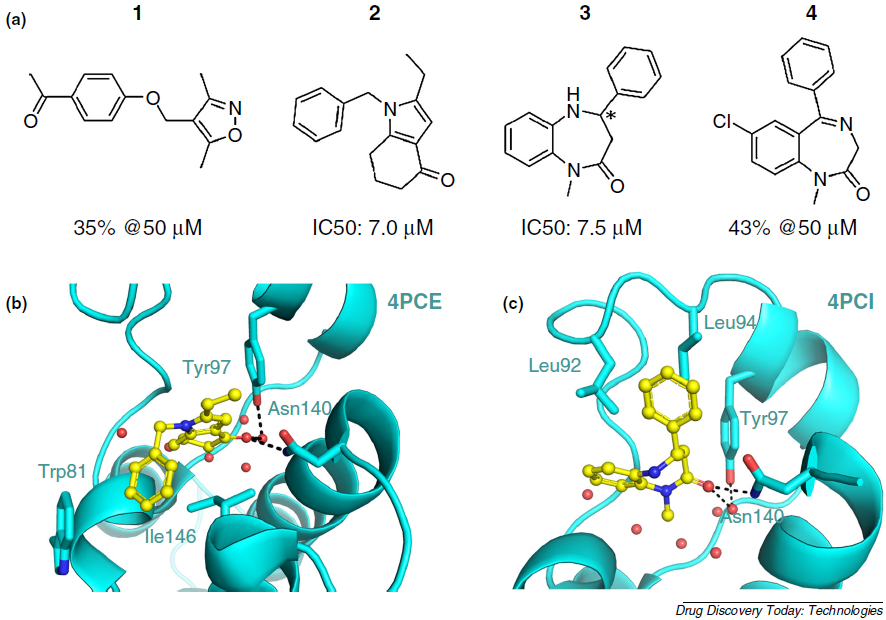
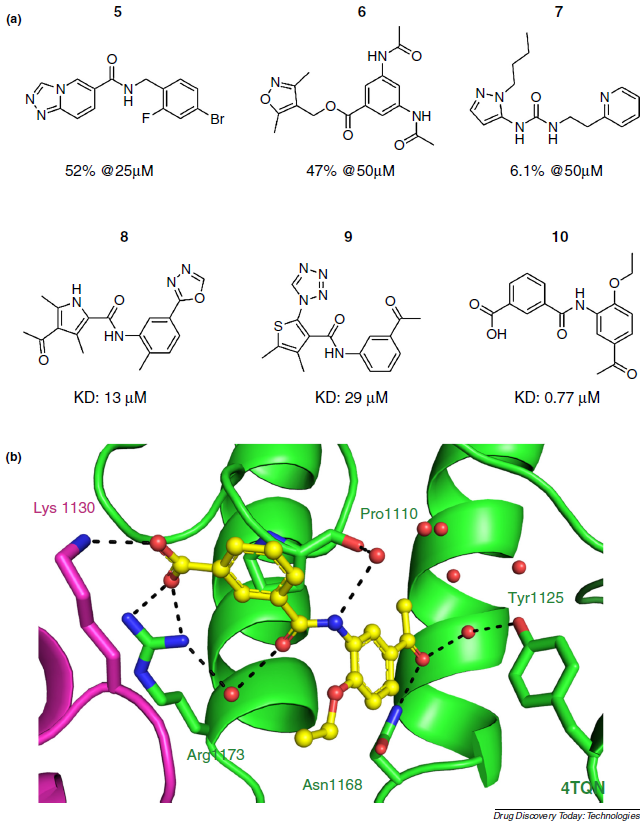
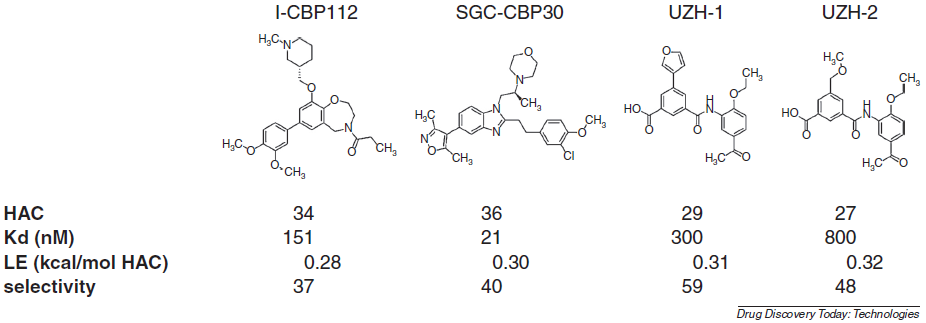
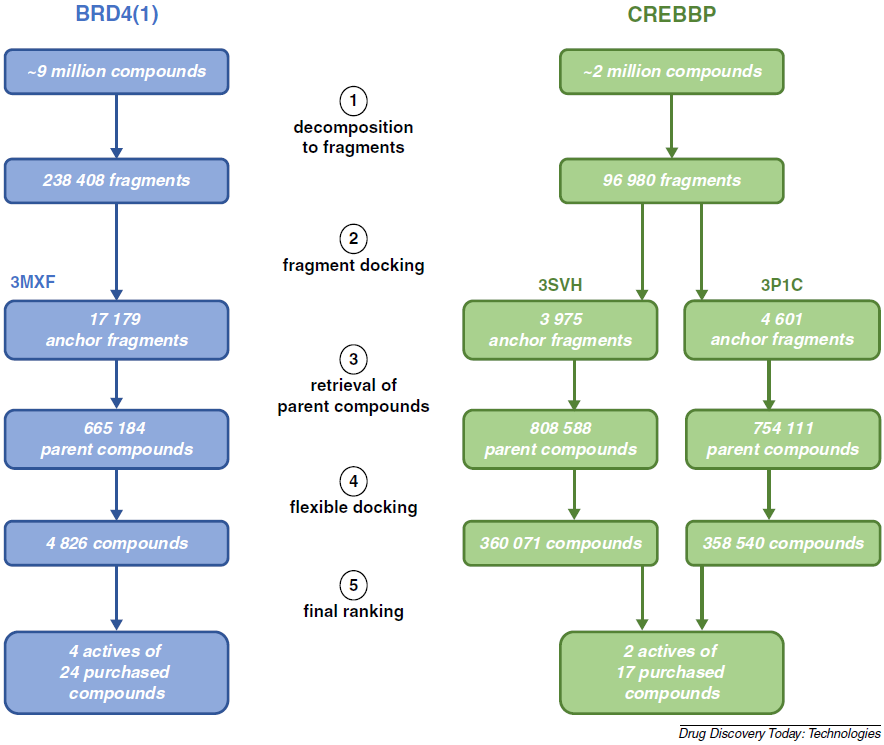
Fragment-based in silico screening of bromodomain ligands
Spiliotopoulos D, Caflisch A
2016. Drug Discov Today Technol, 19:81–90.
DOI: 10.1016/j.ddtec.2016.06.003.
14.
Structural basis for PHDVC5HCHNSD1-C2HRNizp1 interaction: Implications for Sotos syndrome
Berardi A*, Quilici G*, Spiliotopoulos D*, Corral-Rodriguez MA, Martin-Garcia F, Degano M, Tonon G, Ghitti M, Musco G
2016. Nucl Acid Res, 44(7):3448–63. DOI: 10.1093/nar/gkw103.
13.
Fragment-based design of selective nanomolar ligands of the CREBBP bromodomain
Unzue A, Xu M, Dong J., Wiedmer L, Spiliotopoulos D, Caflisch A, Nevado C
2016. J Med Chem, 59(4):1350–6. DOI: 10.1021/acs.jmedchem.5b00172.
12.
Discovery of CREBBP bromodomain inhibitors by high-throughput docking and hit optimization guided by Molecular Dynamics
Xu M, Unzue A, Dong J, Spiliotopoulos D, Nevado C, Caflisch A
2016. J Med Chem, 59(4):1340–9. DOI: 10.1021/acs.jmedchem.5b00171.
11.
GMXPBSA 2.1: A GROMACS tool to perform MM/PBSA and computational alanine scanning
Paissoni C, Spiliotopoulos D, Musco G, Spitaleri A
2015. Comput Phys Commun, 186:105–7. DOI: 10.1016/j.cpc.2014.09.010.
10.
Structural analysis of the binding of type I, I1/2, and II inhibitors to Eph tyrosine kinases
Dong J, Zhao H, Zhou T, Spiliotopoulos D, Rajendran C, Li XD, Huang D, Caflisch A
2014. ACS Med Chem Lett, 6(1):79–83. DOI: 10.1021/ml500355x.
9.
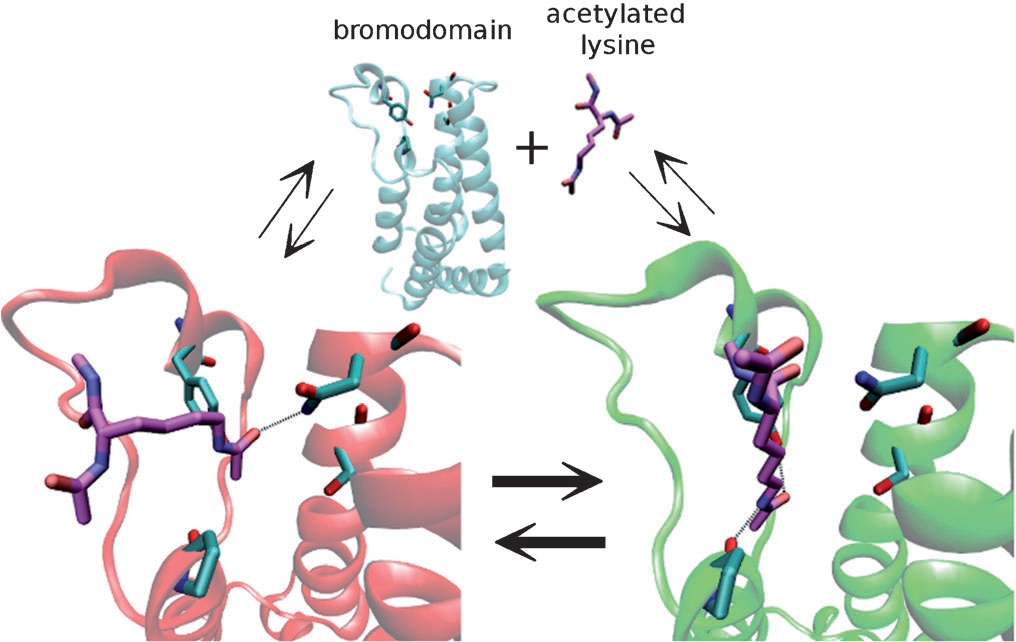
Molecular Dynamics simulations of bromodomains reveal binding‐site flexibility and multiple binding modes
of the natural ligand acetyl‐lysine
Spiliotopoulos D, Caflisch A
2014. Isr J Chem, 54:1084–92. DOI: 10.1002/ijch.201400009.
8.
Discovery of BRD4 bromodomain inhibitors by fragment-based high-throughput docking
Zhao H, Gartenmann L, Dong J, Spiliotopoulos D, Caflisch A
2014. Bioorg Med Chem Lett, 24(11):2493–6.
DOI: 10.1016/j.bmcl.2014.04.017.
7.
GMXPBSA 2.0: A GROMACS tool to perform MM/PBSA and computational alanine scanning
Paissoni C, Spiliotopoulos D, Musco G, Spitaleri A
2014. Comput Phys Commun, 185(11):2920–9.
DOI: 10.1016/j.cpc.2014.06.019.
6.
Sana ME, Spitaleri A, Spiliotopoulos D, Pezzoli L, Preda L, Musco G, Ferrazzi P, Iascone M
2014. Am J Med Genet A, 164(8):2069–73. DOI:
10.1002/ajmg.a.36588.
5.
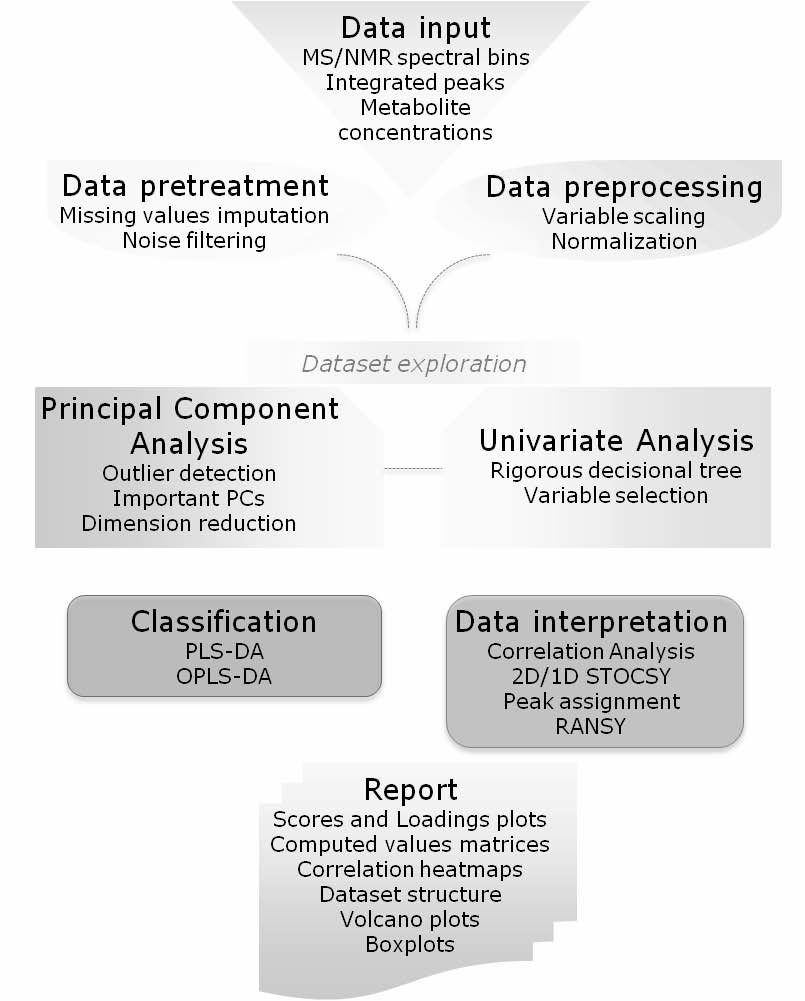
muma, an R package for metabolomics univariate and multivariate statistical analysis
Gaude E, Chignola F, Spiliotopoulos D, García-Manteiga JM, Ghitti M, Spitaleri A, Mari S, Musco G
2013. Current Metabolomics, 1(2):180–9. DOI:
10.2174/2213235X11301020005.
4.
Spiliotopoulos D, Spitaleri A, Musco G
2013. PLoS One, 7(10):e46902. DOI: 10.1371/journal.pone.0046902.
3.
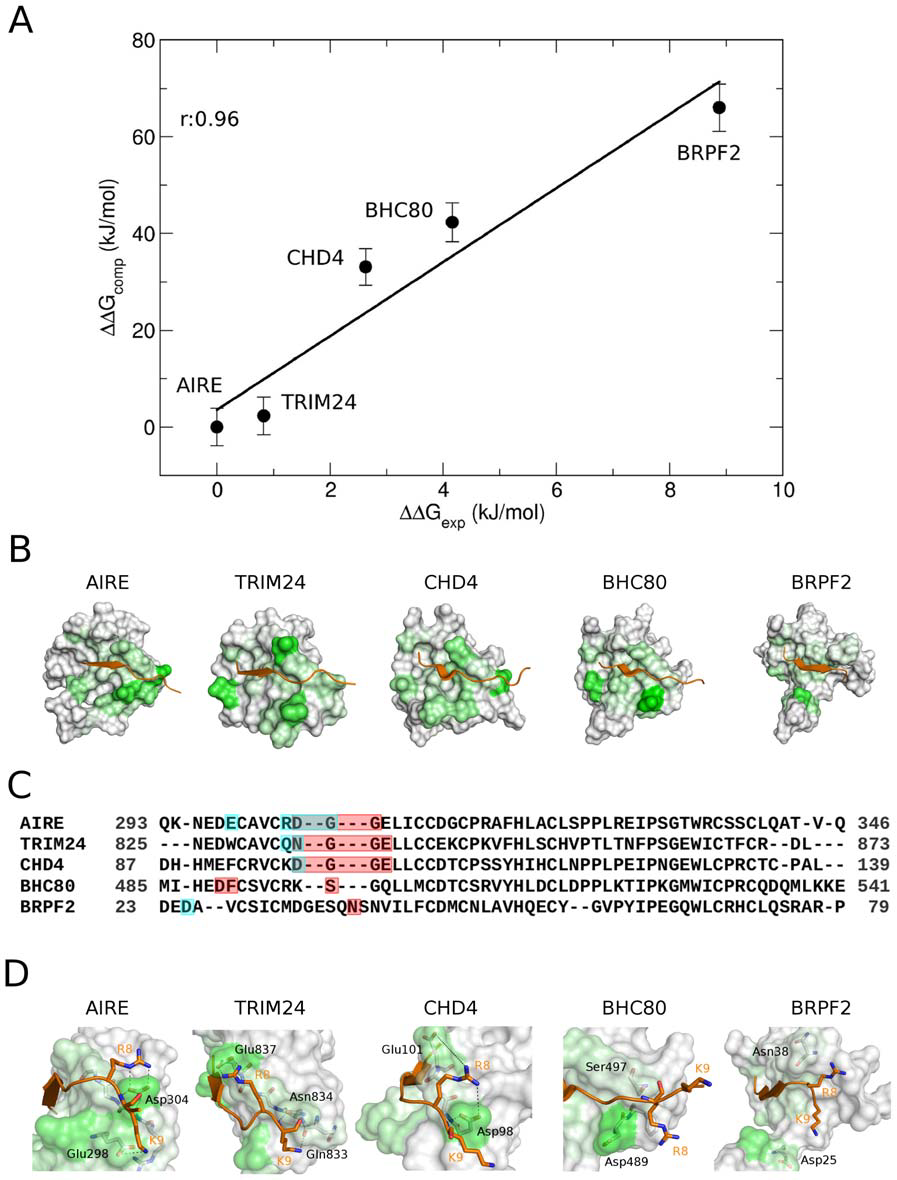
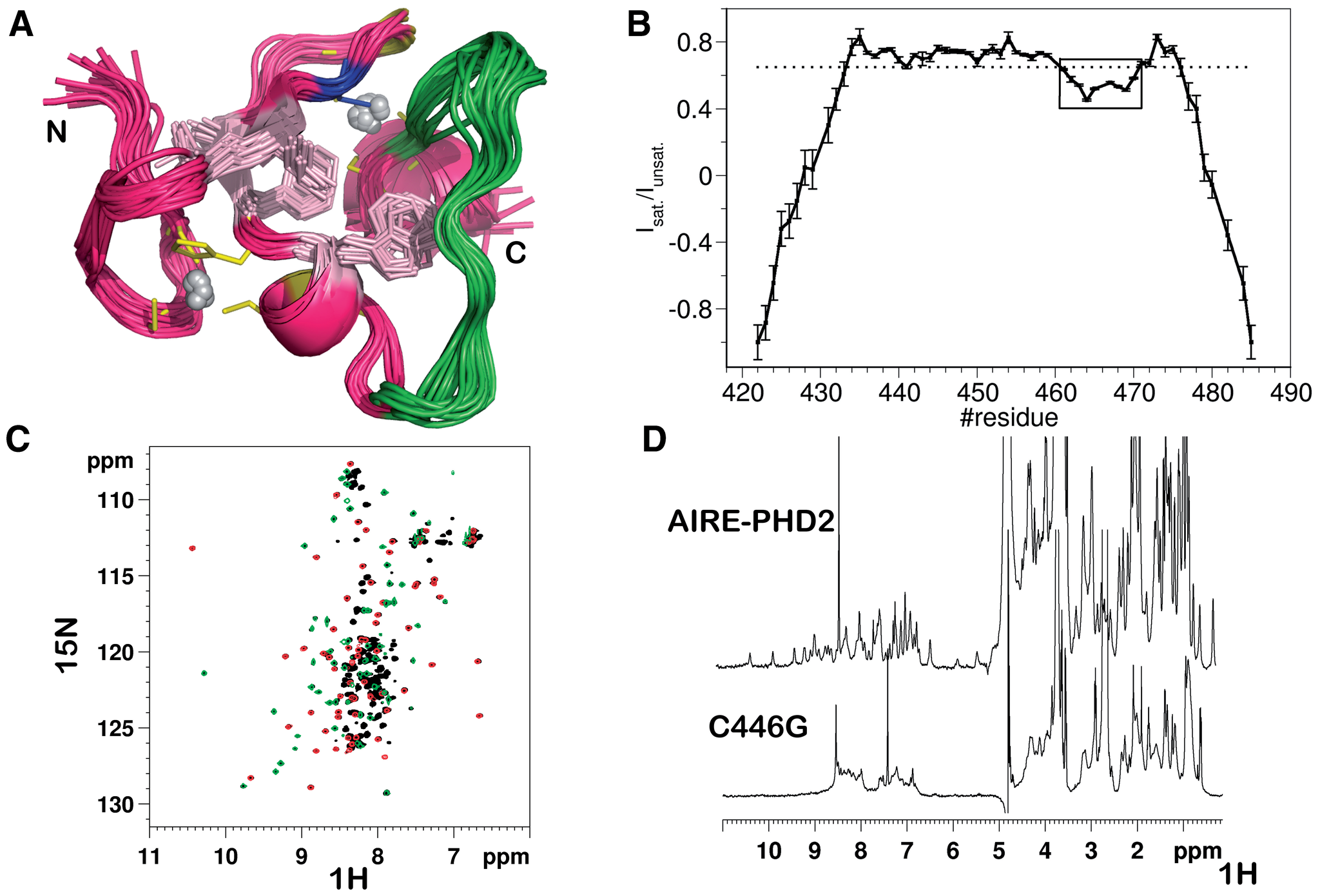
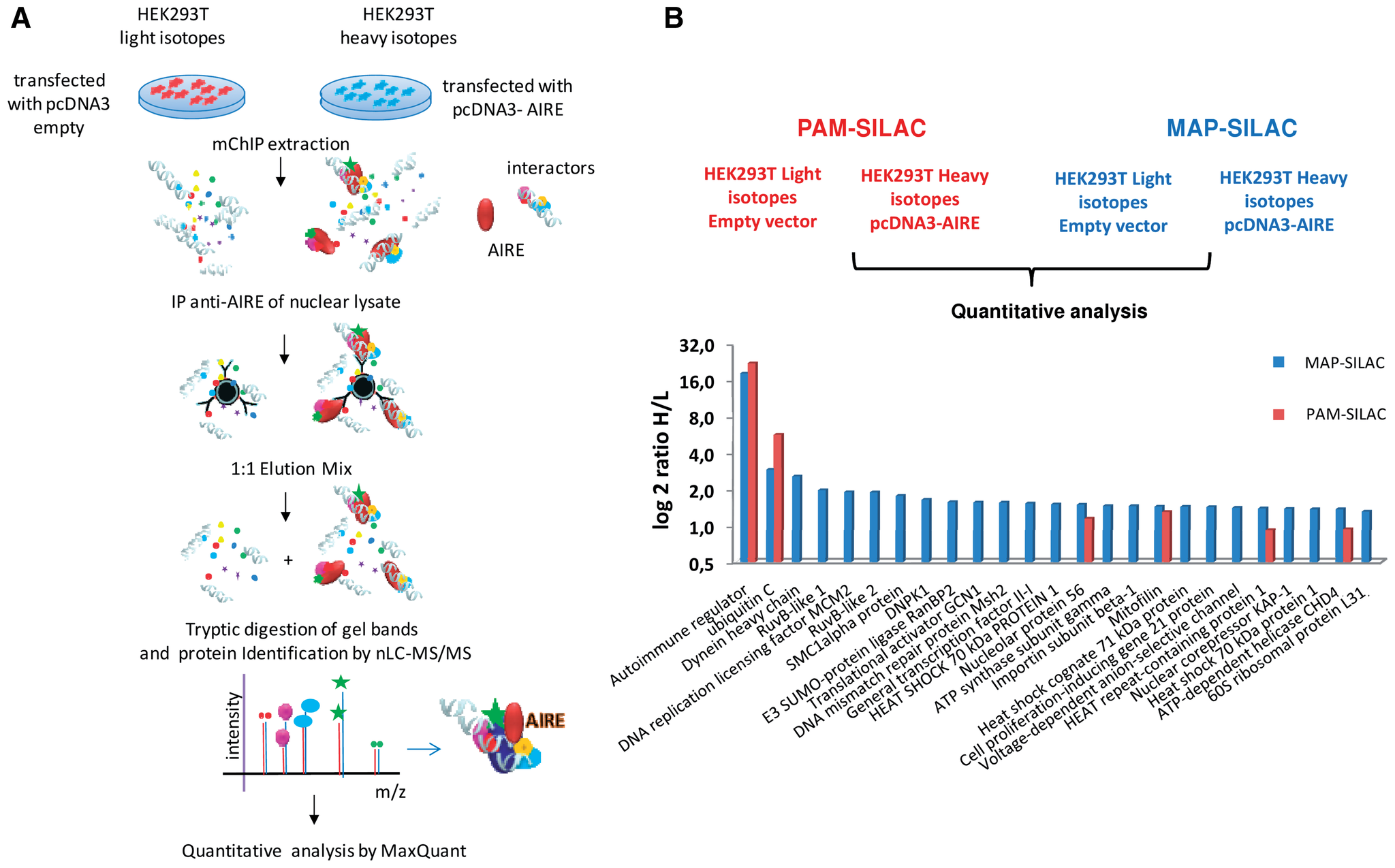
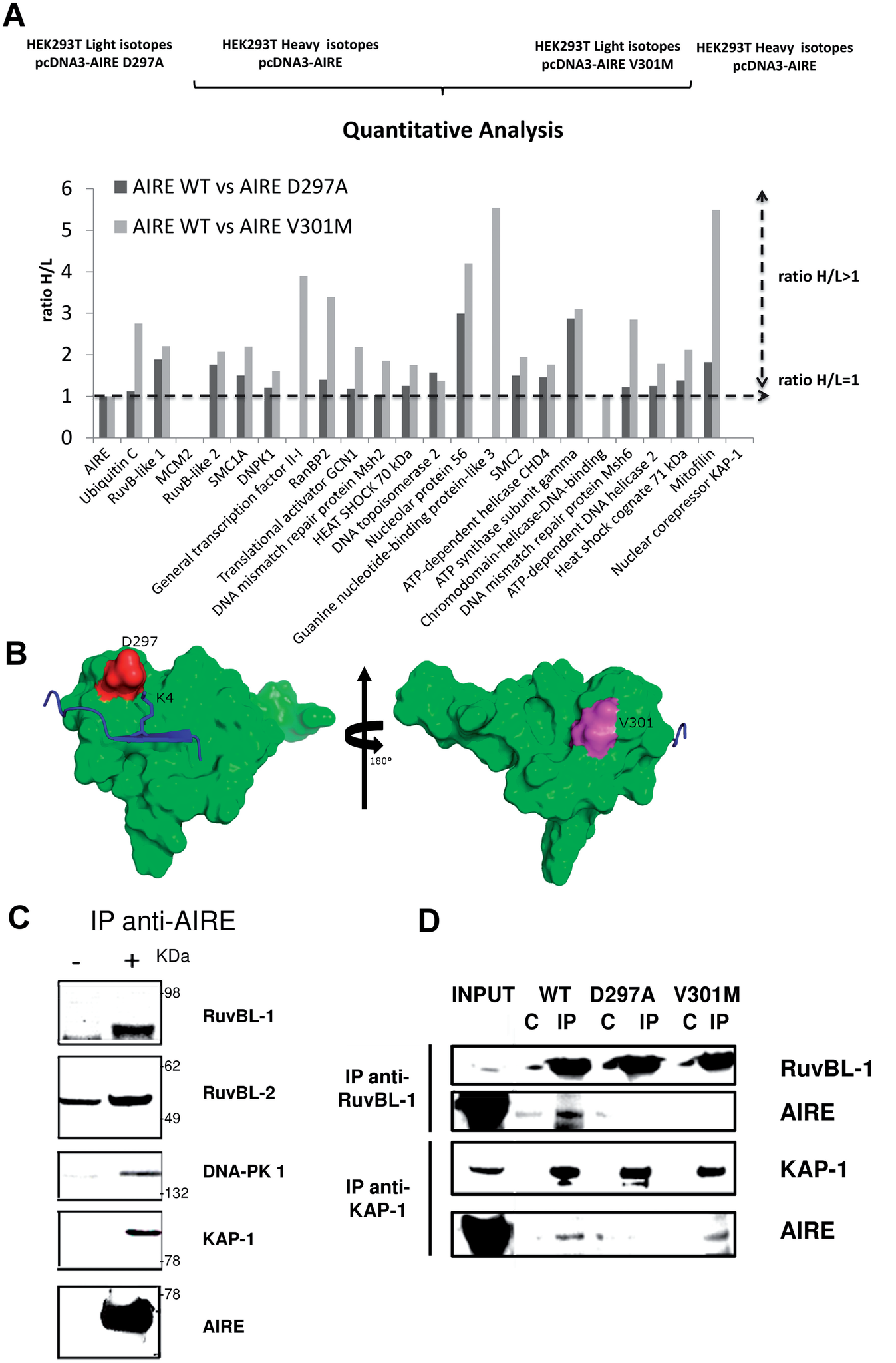
AIRE-PHD fingers are structural hubs to maintain the integrity of chromatin-associated interactome
Gaetani M, Matafora V, Saare M, Spiliotopoulos D, Mollica L, Quilici G, Chignola F, Mannella V, Zucchelli C,
Peterson P, Bachi A, Musco G
2012. Nucl Acid Res, 40(22):11756–68.
DOI: 10.1093/nar/gks933.
2.
A DNA-transposon-based approach to functional screening in Neural Stem cells
Albieri A, Onorati M, Calabrese G, Moiana A, Biasci D, Badaloni A, Camnasio S, Spiliotopoulos D, Ivics Z, Cattaneo
E, Consalez GG
2010. J Biotechnol, 150(1):11–21
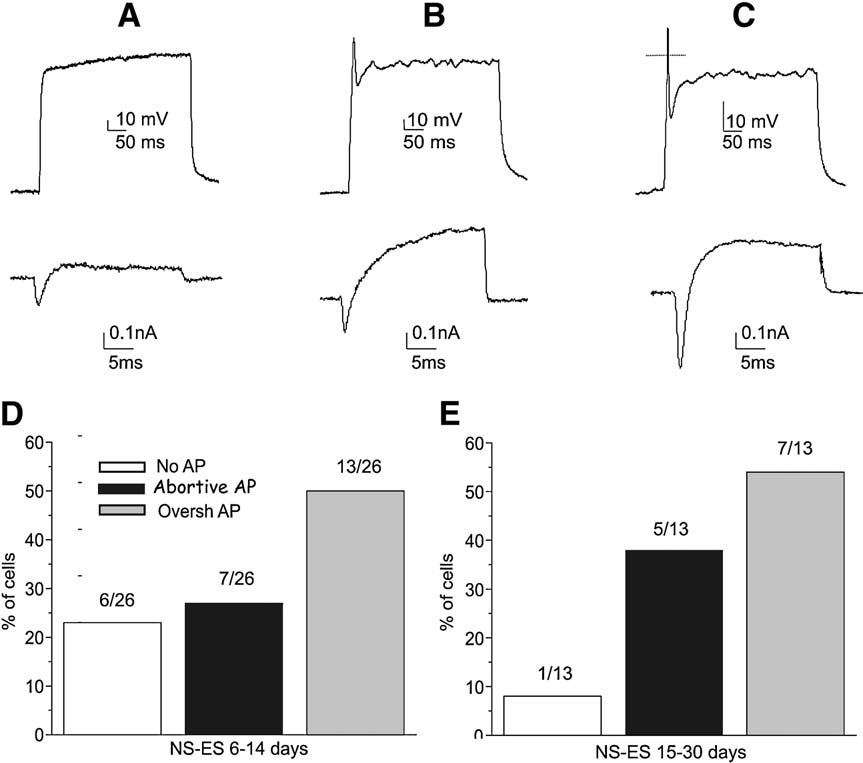
1.
Spiliotopoulos D*, Goffredo D*, Conti L*, Di Febo F, Toselli M, Cattaneo E
2009. Neurobiol Dis, 34(2):320–31. DOI: 10.1016/j.nbd.2009.02.007.
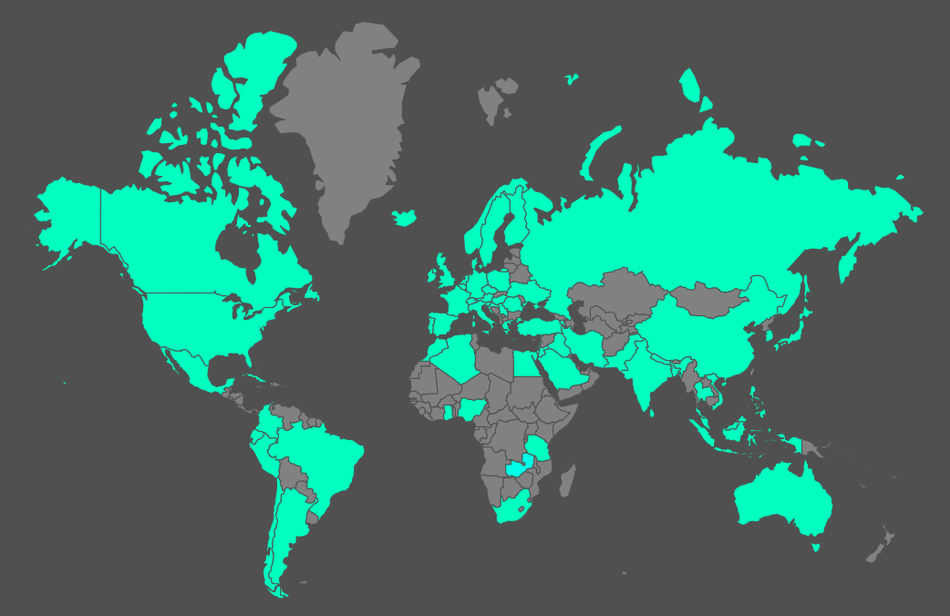
Here's the global localization of the Google searches of my works according to academia.edu (starting Feb 2018).
The works people looked at the most? muma, GMXPBSA 2.0 and GMXPBSA 2.1!
R Packages
Metabolomics Univariate and Multivariate Analysis
Gaude E, Chignola F, Spiliotopoulos D, Mari S, Spitaleri A, Ghitti M
Version: 1.4
License: GPL-2
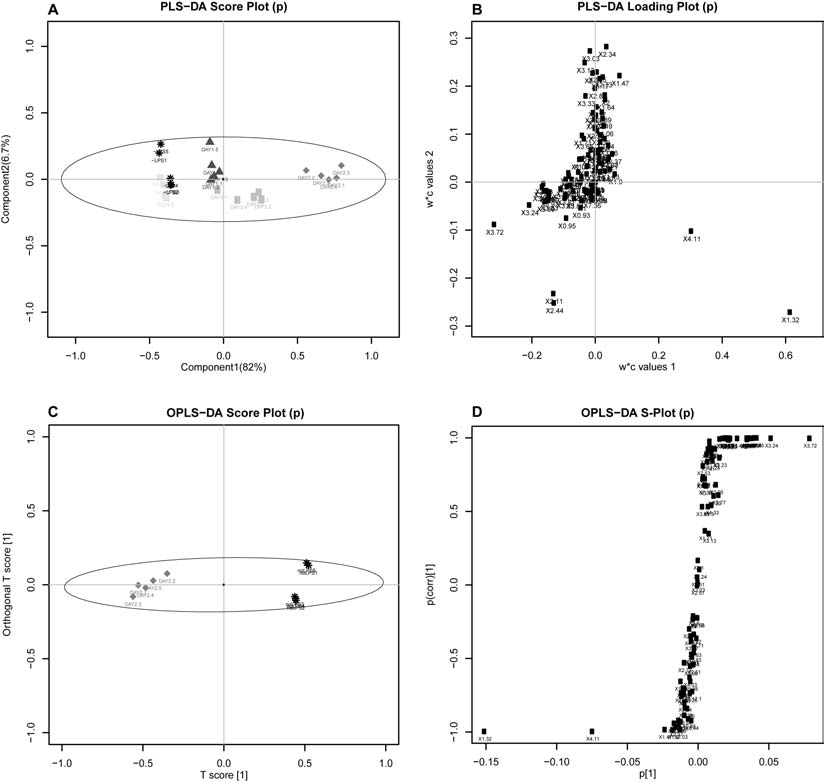
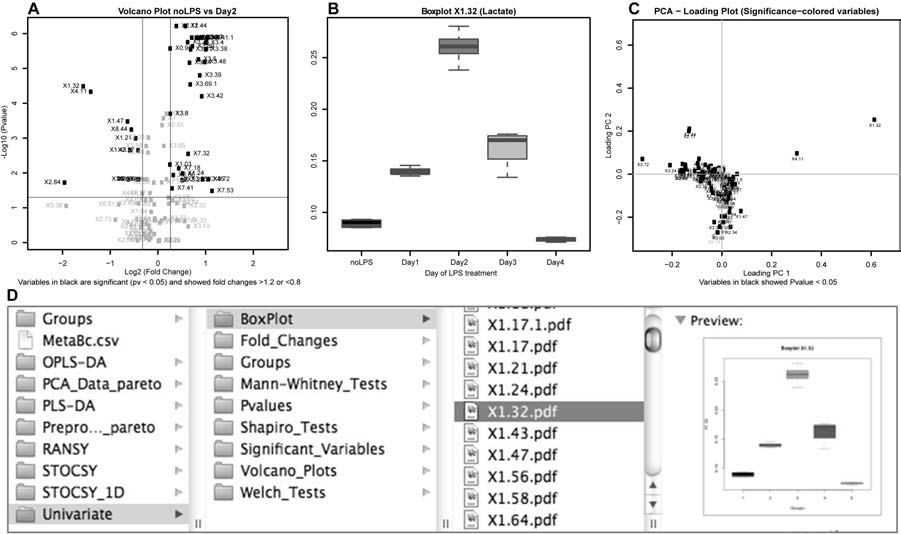
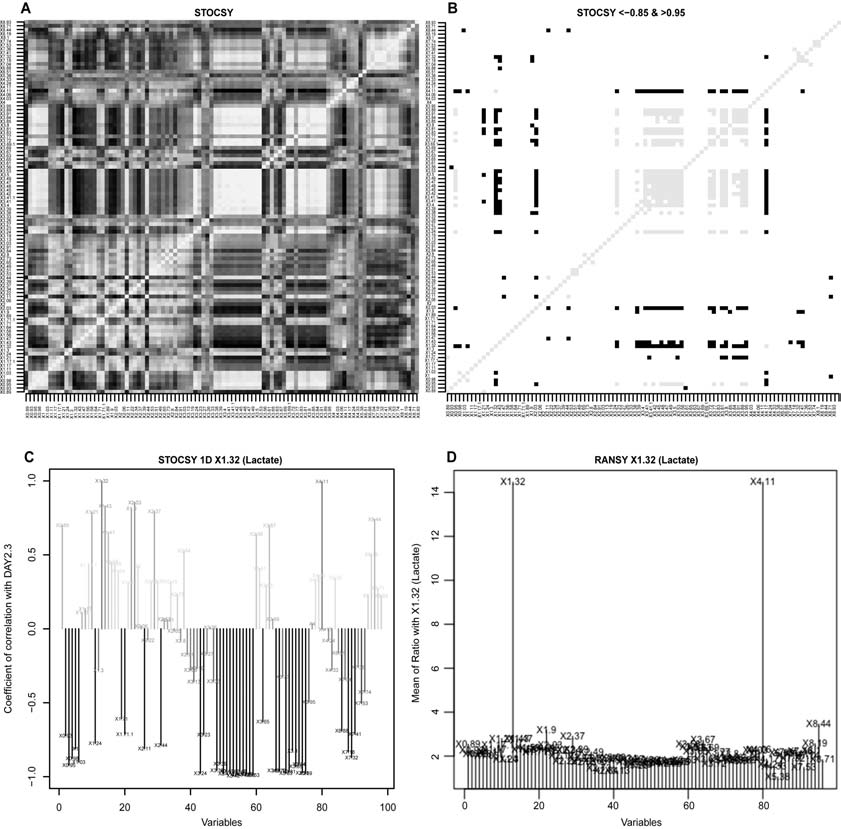
Preprocessing of high-throughput data (normalization and scalings); Principal Component Analysis with help tool for choosing best-separating principal components and automatic testing for outliers; automatic univariate analysis for parametric and non-parametric data, with generation of specific reports (volcano and box plots); partial least square discriminant analysis (PLS-DA); orthogonal partial least square discriminant analysis (OPLS-DA); Statistical Total Correlation Spectroscopy (STOCSY); Ratio Analysis Nuclear Magnetic Resonance (NMR) Spectroscopy (RANSY).
Other papers
3.
Hothorn L, Spiliotopoulos D, Marin-Kuan M, Ritz C
zenodo, uploaded September 1st 2021. DOI:10.5281/zenodo.5356885.
2.
Similarity of multiple dose-response curves in interlaboratory studies in regulatory toxicology
Hothorn L, Spiliotopoulos D
arXiv, submitted September 25th 2020.
1.
Molecular and computational analysis of regulation of hSos1, the major activator of the proto-oncoprotein Ras
Farina M, Sacco E, Greco C, Busti S, Spiliotopoulos D, De Gioia L, Liberati D, Alberghina L, Vanoni
M
International Congress ECOBIOSYS 2009 Classification and Forecasting Models, Milan.
GMXPBSA in the world

Dimitrios Spiliotopoulos's website -- still in preparation!
Everything you see on this website is still under construction, so feel free to let me know what you think! ;-)
To contact me check the "Contacts & Links" page!